C A ↪D C ↪EBVBbc*BVBdd->C BVBda->D BVBc_->E
A Model of Gene Regulation
The regulatory regions in DNA play an key role in determining when genes are expressed. These regions of DNA contain binding sites for regulatory proteins. How these binding sites are arranged, and the affinity of regulatory proteins for these sites determines, in part, whether particular genes are expressed or not.
I say DNA encodes rules for three reasons:
- The expression of a gene is influenced by a combination of the presence of regulatory proteins and other cellular conditions, such as the presence of certain ligands or co-factors.
- Modification to the regulatory region can alter these conditions.
- The alterations of these regions has a fine-grained, range of different modifications Something about open-ended, many possibilities, fine-grained.
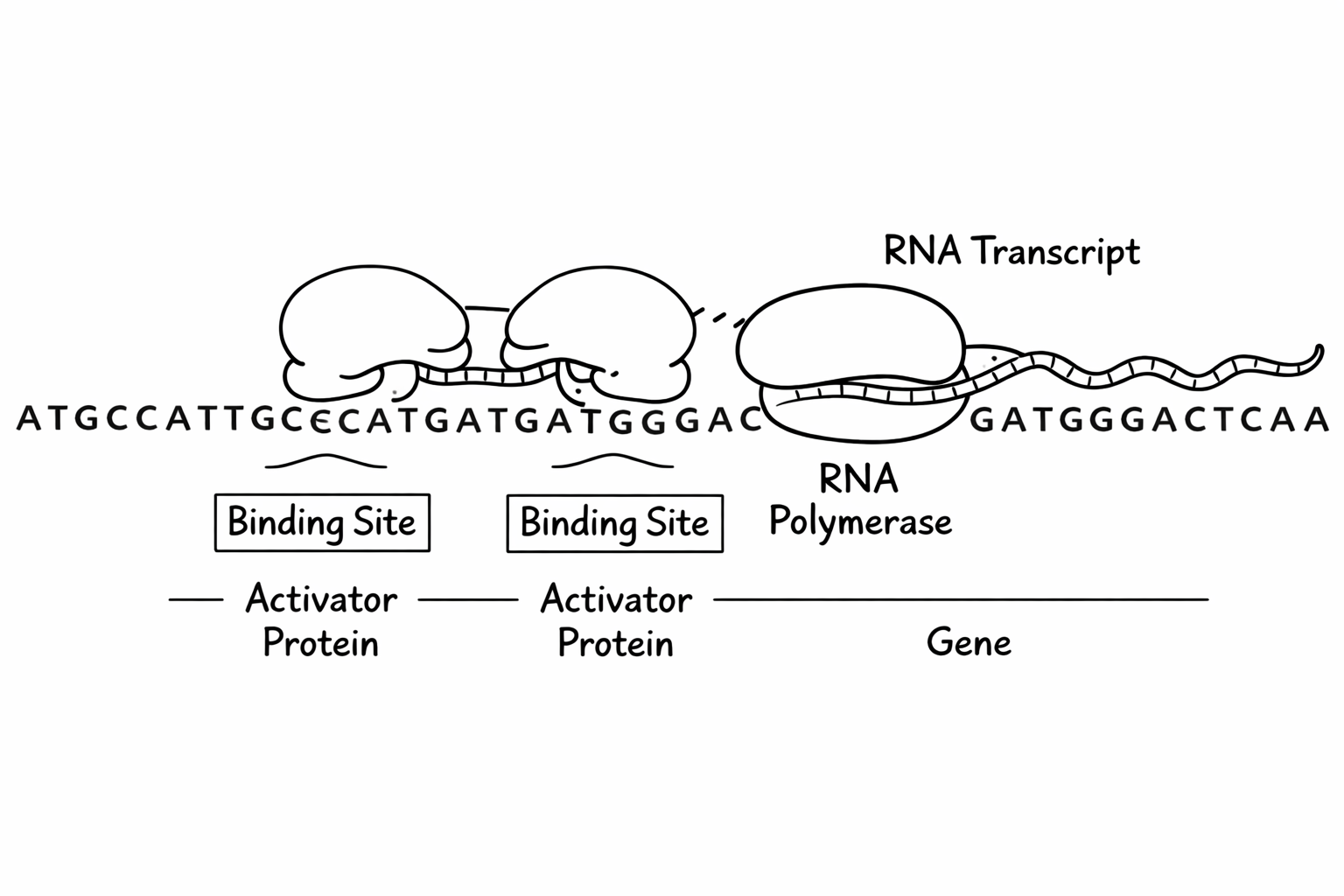
The AND programming language captures a simple model of how these regions work and change over time. - The program state represents the conditions in the cell that influence gene -expression. - The syntax of AND expressions mimics how these rules are encoded in the DNA. - The semantics of the AND expressions is captures how these region influence gene expression.
Binding and transcription in AND
We can treat the program from the previous section like a stretch of DNA with regulatory regions and genes.
Each expression in an AND program represents a gene. The target register of the expression represents the protein produced when the gene is expressed. When a gene is switched on, that particular protein will be present in the next time step.
Structural genes produce proteins that directly affect the organism’s traits, while regulatory genes produce proteins that influence the expression of other genes. We distinguish the type of gene by the type of register it writes to:
- An expression that writes to an output register is a structural gene.
- An expression that writes to a memory register is a regulatory gene.
The activation condition of an expression represents the regulatory region of the gene. This regulatory region encodes the conditions under which the gene is expressed, and thus when the protein is produced.
We can interpret the components of the condition in biological terms:
- The lowercase register names in the condition are regulatory motifs that bind to the regulatory proteins or introduced elements that are present:
Abinds to motifa,Bbinds to motifb, and so on. - The operators, such as
BVBcaptures describe how the two proteins binding at their motifs interact. - The combiners,
*and+, represent how multiple regulatory modules can either act cooperatively (*) or independently (+) to regulate gene expression.
Program state. The conditions in the cell at each time are capture by the values of the registers in the program state.
Input registers are a way to capture additional conditions in the cell that are not the direct result of genes expression, such as the presence ligands or co-factors, that influence gene expression. In reality, external factors influence gene expression by interacting with existing proteins. Here, we simplify this by allowing these factors to directly “bind” to the regulatory region.
Running the program
Lets do this.